Why Alt-R A.s. Cas12a (Cpf1) Ultra?
CRISPR-Cas12a (Cpf1) is an RNA-guided DNA endonuclease that is an alternative to the commonly used Streptococcus pyogenes Cas9 (S.p. Cas9) enzyme [1]. Unlike S.p. Cas9, which recognizes NGG PAM sequences [2, 3], Cas12a recognizes TTTV (V = A/G/C) PAM sites, thereby permitting genome editing in organisms with AT-rich genomes. A.s. Cas12a is an attractive option for genome editing applications due to its AT-rich PAM sequence [1], its highly specific DNA recognition and cleavage mechanism [4, 5], and its native reliance on a single, short guide RNA. However, comparison studies showed that the wild-type A.s. Cas12a enzyme lacks the potency and site-to-site reliability of S.p. Cas9 when used with relatively GC-rich genomes such as the human genome. Thus, we undertook a directed evolution approach to improve on the performance of the wild-type enzyme. In this article, we describe our method of developing Alt‑R A.s. Cas12a (Cpf1) Ultra, its cleavage activity, and its performance with TTTV PAM site selection. In addition, we demonstrate that A.s. Cas12a Ultra exhibits increased editing efficiency at TTTN (N = A/G/C/T) PAM sites across different cell lines, outperforms both wild-type A.s. and L.b. Cas12a proteins at 30 and 37°C, and significantly increases homology-directed repair (HDR) rates.
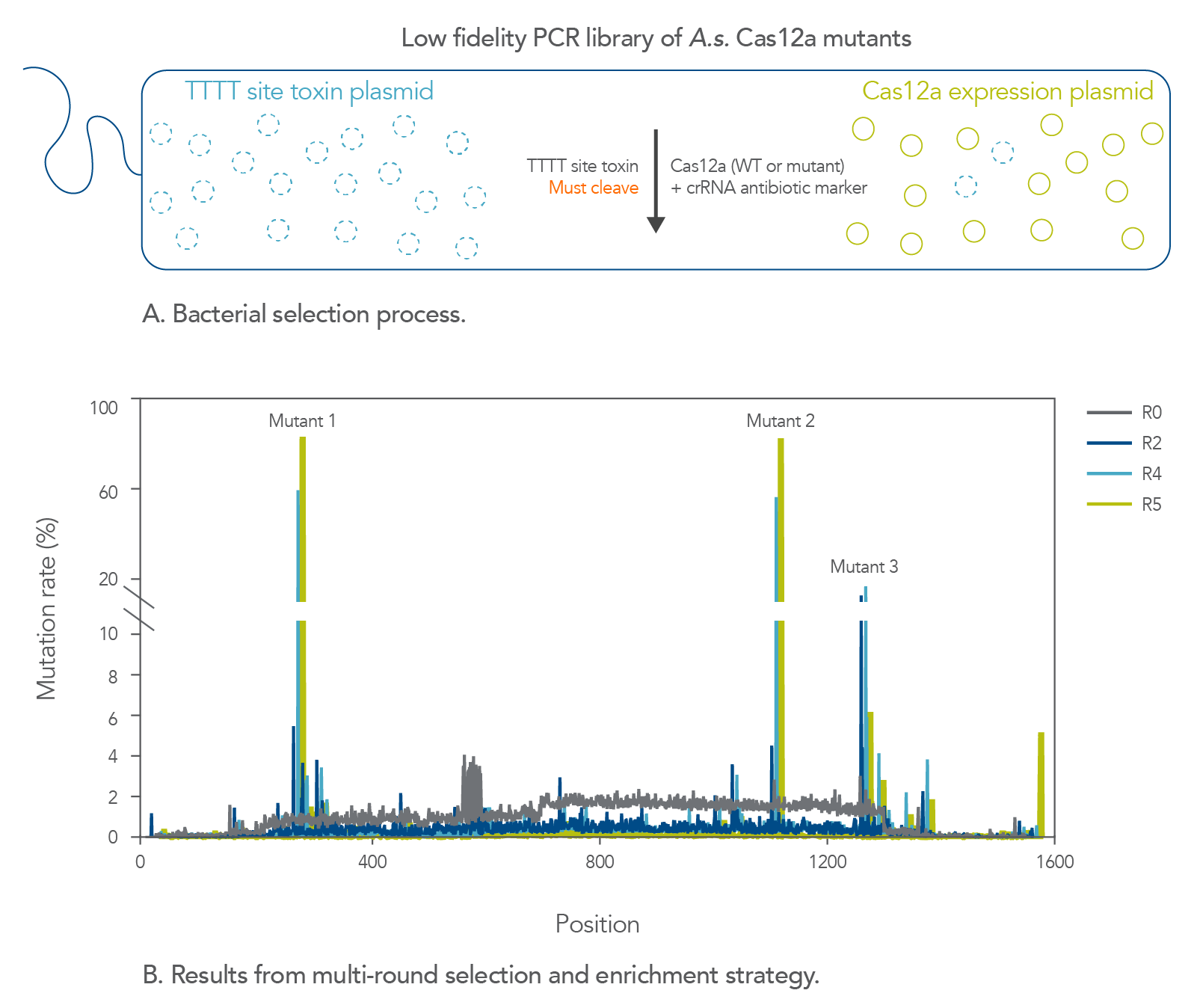
Isolation of an A.s. Cas12a mutant with increased cleavage activity
CRISPR enzymes function natively in bacteria as part of a primitive immune system that helps to maintain genomic integrity from invading bacteriophages [6]. The finding that these enzymes, which are simply RNA-targeted DNA endonucleases, could introduce targeted dsDNA breaks in human cells was quite remarkable [3] . Nevertheless, these enzymes did not evolve to work in the mammalian nucleus and are not always perfectly adapted to function in that environment. S.p. Cas9, for example, can facilitate an alarming amount of unwanted off-target editing when targeting certain genomic sites [7]. This finding is perhaps not surprising given that eukaryotic genomes are generally far larger than those of prokaryotes, and thus S.p. Cas9 did not evolve to target a single loci while editing the human genome. At IDT, we have given nature a boost by applying directed evolution principles to both S.p. Cas9 and A.s. Cas12a to make them better suited for genome editing in eukaryotic cells.
Previously, to address the low fidelity of S.p. Cas9, we engineered the Alt-R HiFi Cas9 protein, which is a mutant form of S.p. Cas9 that was evolved to decrease its off-target editing activity [8]. While fidelity (i.e., site-specific targeting as opposed to targeting and cleavage of unintended sites) was our main concern with S.p. Cas9, the principal issue we encountered with A.s. Cas12a was poor site-to-site consistency in terms of on-target editing efficiency. We therefore set out improve the reliability of A.s. Cas12a without compromising its lower off-target activity and AT-rich PAM site preference. We developed a bacterial-based selection assay to select for A.s. Cas12a mutants that demonstrate increased cleavage activity (Figure 1). We selected A.s. Cas12a variants with cleavage efficiencies that are higher than those observed with the wild-type enzyme at non-canonical TTTT PAM sites for two reasons. First, this specific motif is significantly more prevalent throughout the human genome than the other three TTTV motifs. Second, A.s. Cas12a generally recognizes TTTT PAM sequences poorly. After multiple rounds of selections, we successfully enriched several mutations, the best of which ultimately made up the Alt-R A.s. Cas12a Ultra protein.
Alt-R A.s. Cas12a Ultra dramatically increases editing efficiency and HDR at TTTN PAM sites
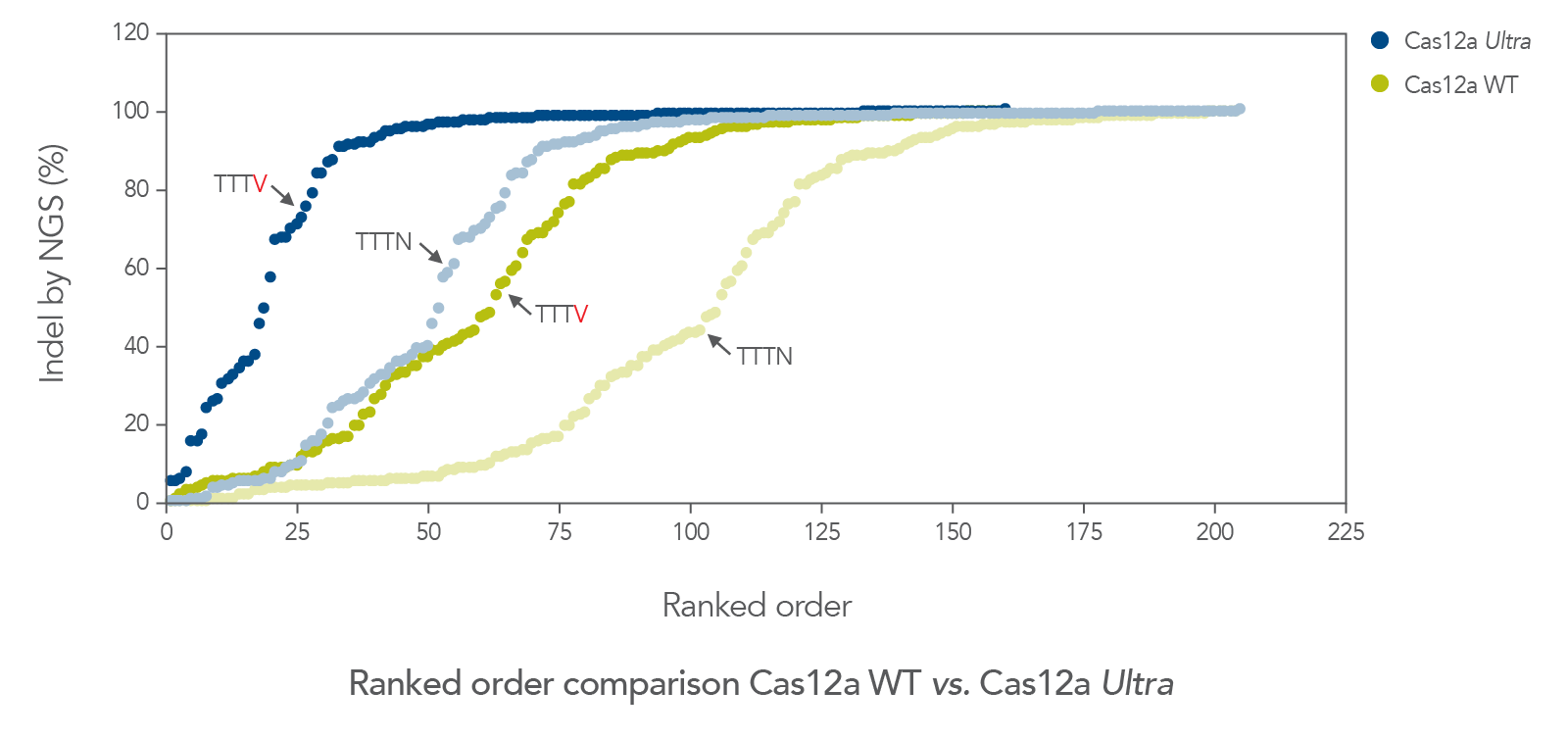
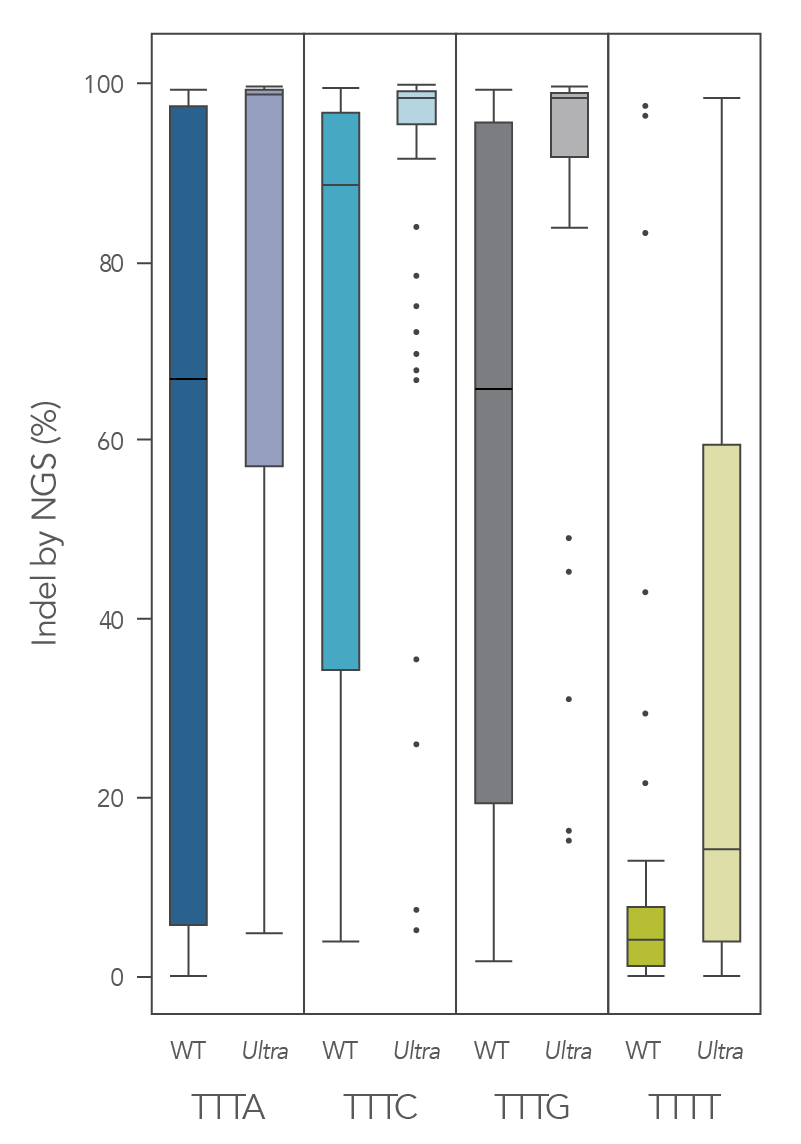
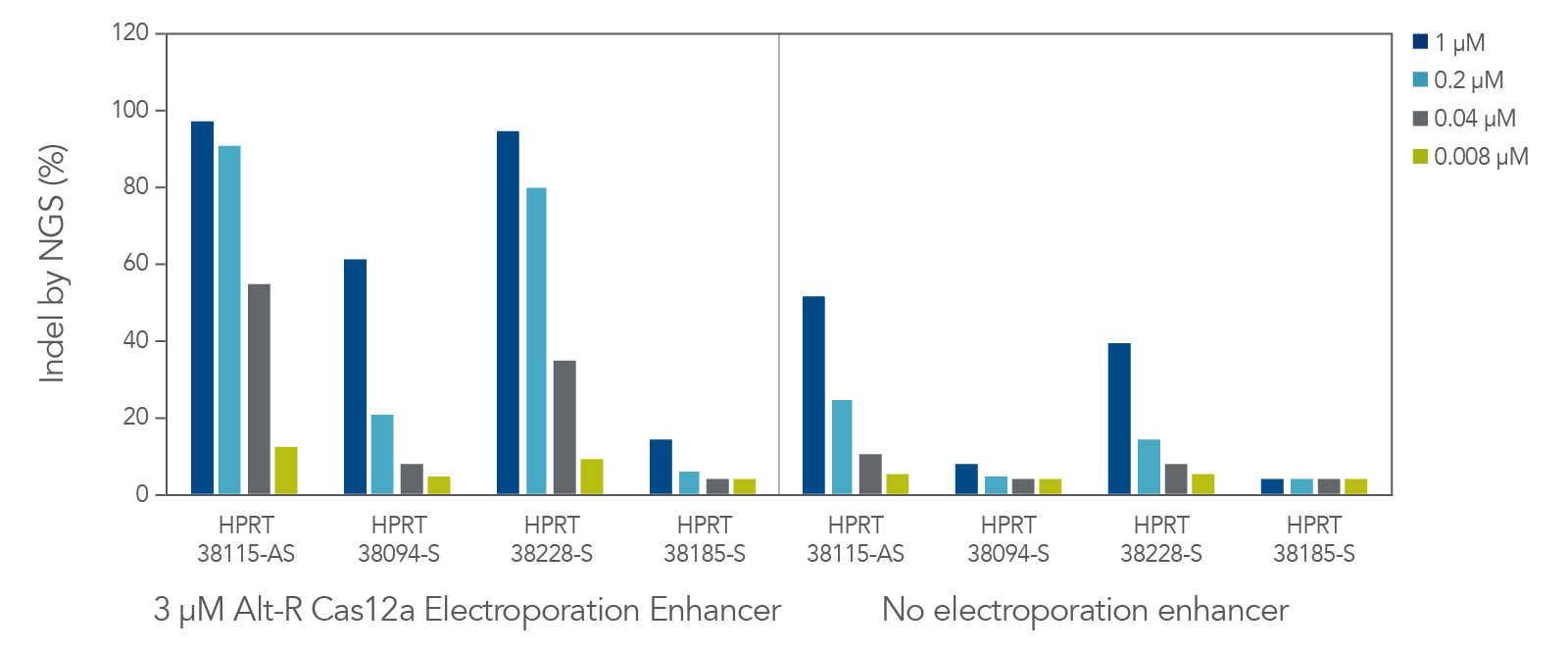
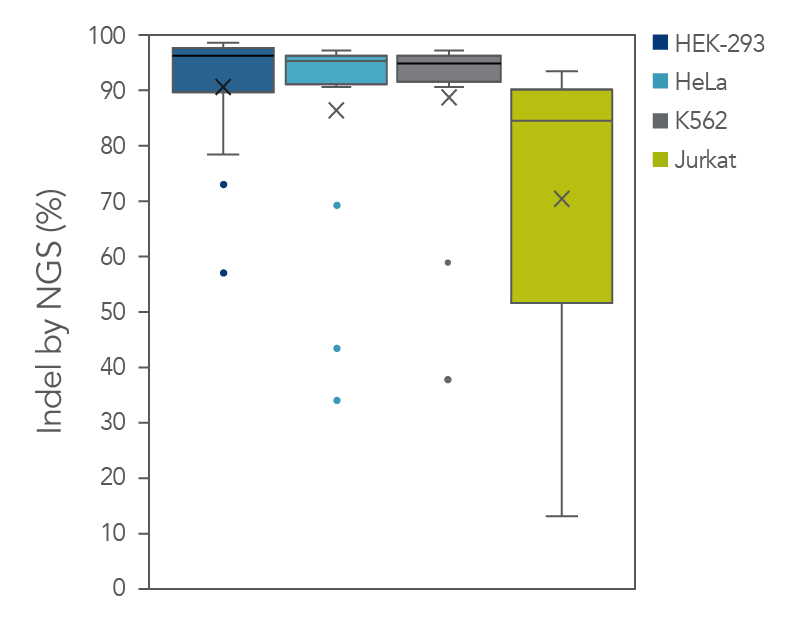
The cleavage efficiency of A.s. Cas12a Ultra was compared to that of the wild-type Cas12a V3 enzyme at both TTTV and TTTN PAM sites when delivered into human cells by electroporation as ribonucleoprotein (RNP) complexes. We examined 216 guides in HEK-293 cells (96 sites) and Jurkat cells (120 sites). For the Cas12a Ultra nuclease with TTTV PAM selection, the average editing efficiency was 88% (indels, determined by NGS), and the median editing efficiency was 99% (Figure 2). These data were further examined to analyze the performance at TTTA, TTTC, TTTG, and TTTT PAM sites. Remarkably, the Cas12a Ultra nuclease dramatically increased performance at all TTTN PAMs, including notoriously difficult-to-edit TTTT PAM sites (Figure 3). Importantly, the Alt-R Cas12a (Cpf1) Electroporation Enhancer was co-delivered in all these experiments. As we had previously shown with wild-type A.s. Cas12a V3 nuclease, we now found that the Cas12a Electroporation Enhancer also dramatically increases the performance of A.s. Cas12a Ultra nuclease (Figure 4). The Cas12a Ultra nuclease was examined in multiple different human cell lines and performed well in all cases (Figure 5).
n = 3
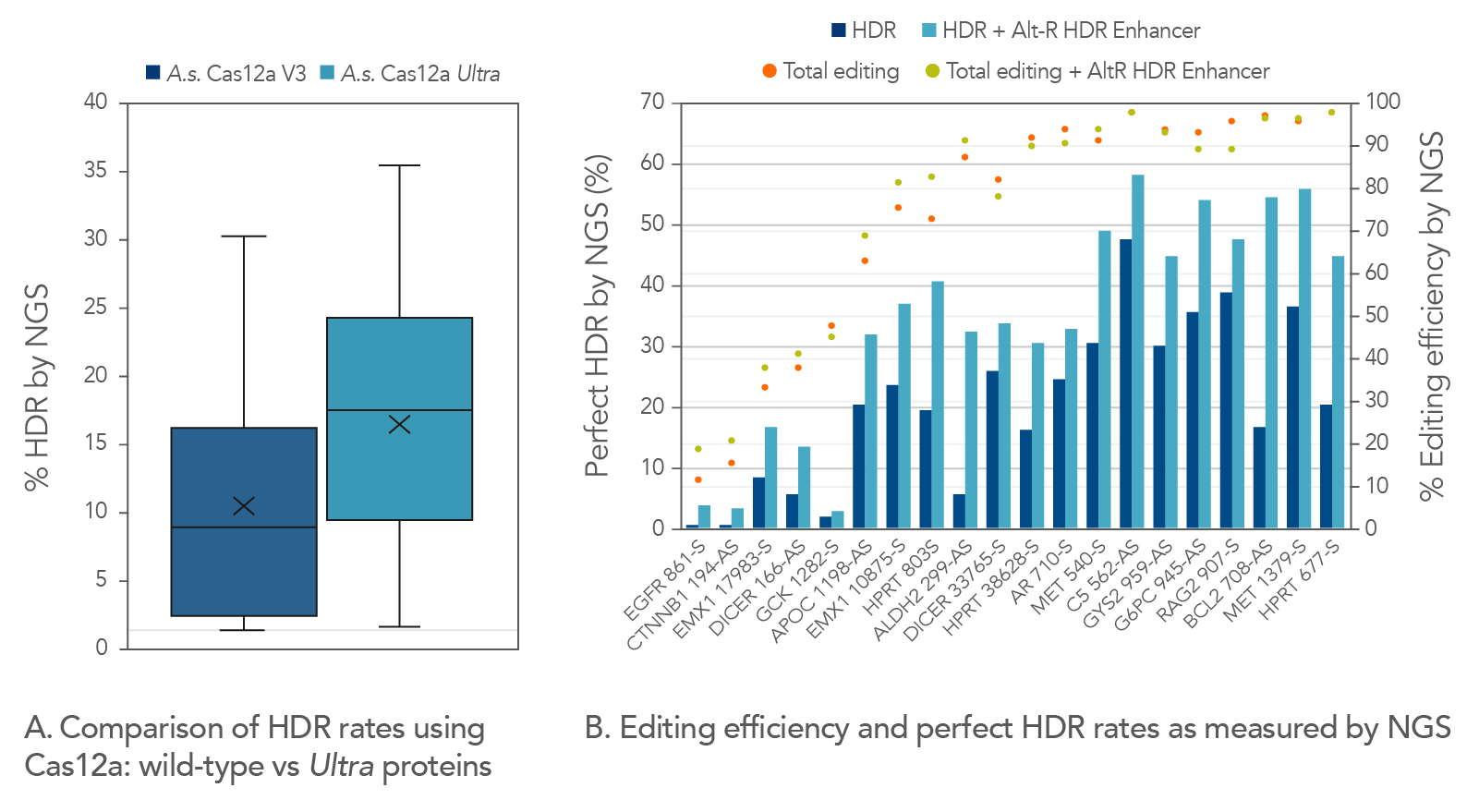
Alt-R A.s. Cas12a Ultra is a universal Cas12a solution that increases overall HDR rates
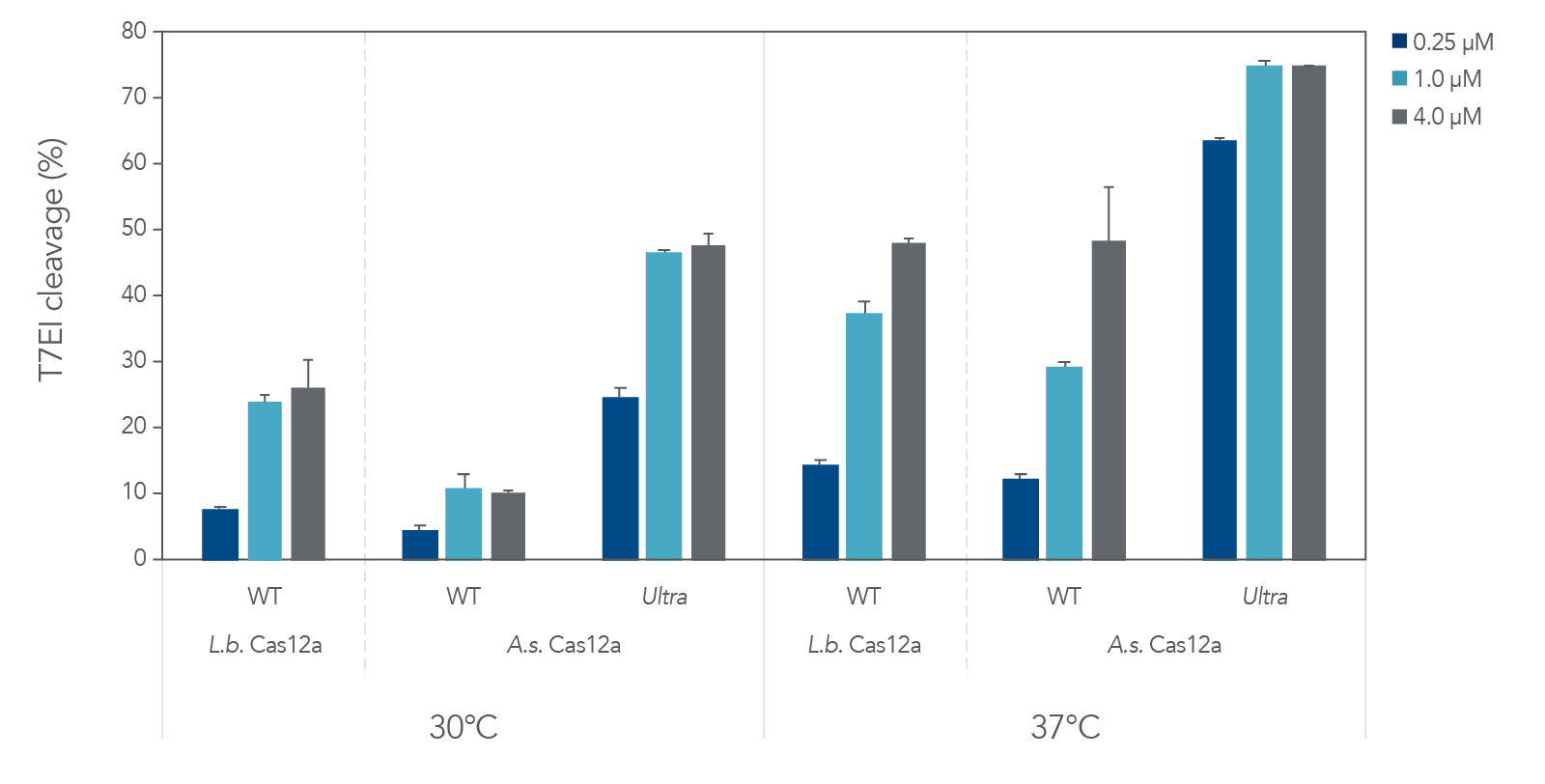
A.s. Cas12a protein, in the wild-type form, performed poorly at low temperatures (25–32ºC) required for delivery into ectothermic organisms such as plant species. In these cases, the Lachnospiraceae bacterium variant L.b. Cas12a has typically been used, as this protein mitigates efficient editing at low temperatures. We compared editing efficiencies in live cells at both 30 and 37ºC, and we found that the A.s. Cas12a Ultra nuclease dramatically outperforms both wild-type A.s. Cas12a V3 and L.b. Cas12a proteins at all temperatures and is a universal Cas12a solution to achieve high editing efficiencies (Figure 6). Finally, we examined the ability of A.s. Cas12a Ultra nuclease to facilitate high levels of HDR in Jurkat cells, typically known for low rates of HDR.
We compared Cas12a Ultra-dependent HDR rates (analyzed by NGS) to those obtained with the wild-type A.s. Cas12a V3 nuclease, and we found that the Cas12a Ultra nuclease approximately doubled HDR rates when using Ultramer™ ssDNA oligonucleotide donor templates (Figure 7).
- Alt-R A.s. Cas12a Ultra nuclease dramatically increases editing efficiency, making the CRISPR-Cas12a system as useful and reliable as the S.p. Cas9 system.
- TTTV PAM site selection, in combination with Alt-R Cas12a Electroporation Enhancer, typically results in greater than 90% editing efficiency with most guides.
- Alt-R A.s. Cas12a Ultra nuclease is a universal Cas12a solution.
- HDR rates are often doubled with the use of Alt-R A.s. Cas12a Ultra nuclease.

