rhAmp™ SNP Assays
Genotyping assays with flexible design and consistent quality
rhAmp SNP Assays offer a collection of predesigned sequences containing >10 million human SNPs, including a broad selection of functionally tested ADME SNP assays. A custom assay design pipeline is also available for newly discovered human SNPs or assay designs for other species.
Ordering
rhAmp SNP Assays are a core component of a complete genotyping solution that offers precision, reliability, and a wide variety of design options. Choose your target SNPs from a predesigned library containing >10 million SNPs, including 330,000 common human SNPs (minor allele frequencies >1%) found in RefSeq. Custom assays for other human SNPs or SNPs in other species can also be created using the rhAmp Genotyping Design Tool. Use rhAmp SNP Assays with rhAmp Genotyping Master Mix and rhAmp Reporter Mix (with or without reference dye) to form a complete amplification and reporting reaction chemistry.
- Generate reliable data with >99.5% call accuracy for over 90% of assays tested
- Test markers efficiently using our small pack size
- Interrogate SNPs in difficult sequence regions with amplicon sizes as small as 40 bp
- Gain confidence in your data by using gBlocks™ Gene Fragments as control templates
The rhAmp SNP Genotyping System is a complete solution requiring the following 3 components:
- rhAmp SNP Assays, or rhAmp ADME SNP Assays
- rhAmp Genotyping Master Mix, required for activation and PCR amplification of all rhAmp SNP Assays
- rhAmp Reporter Mix, available with or without reference dye for compatibility with all common qPCR instruments
Product Details
rhAmp SNP Genotyping technology
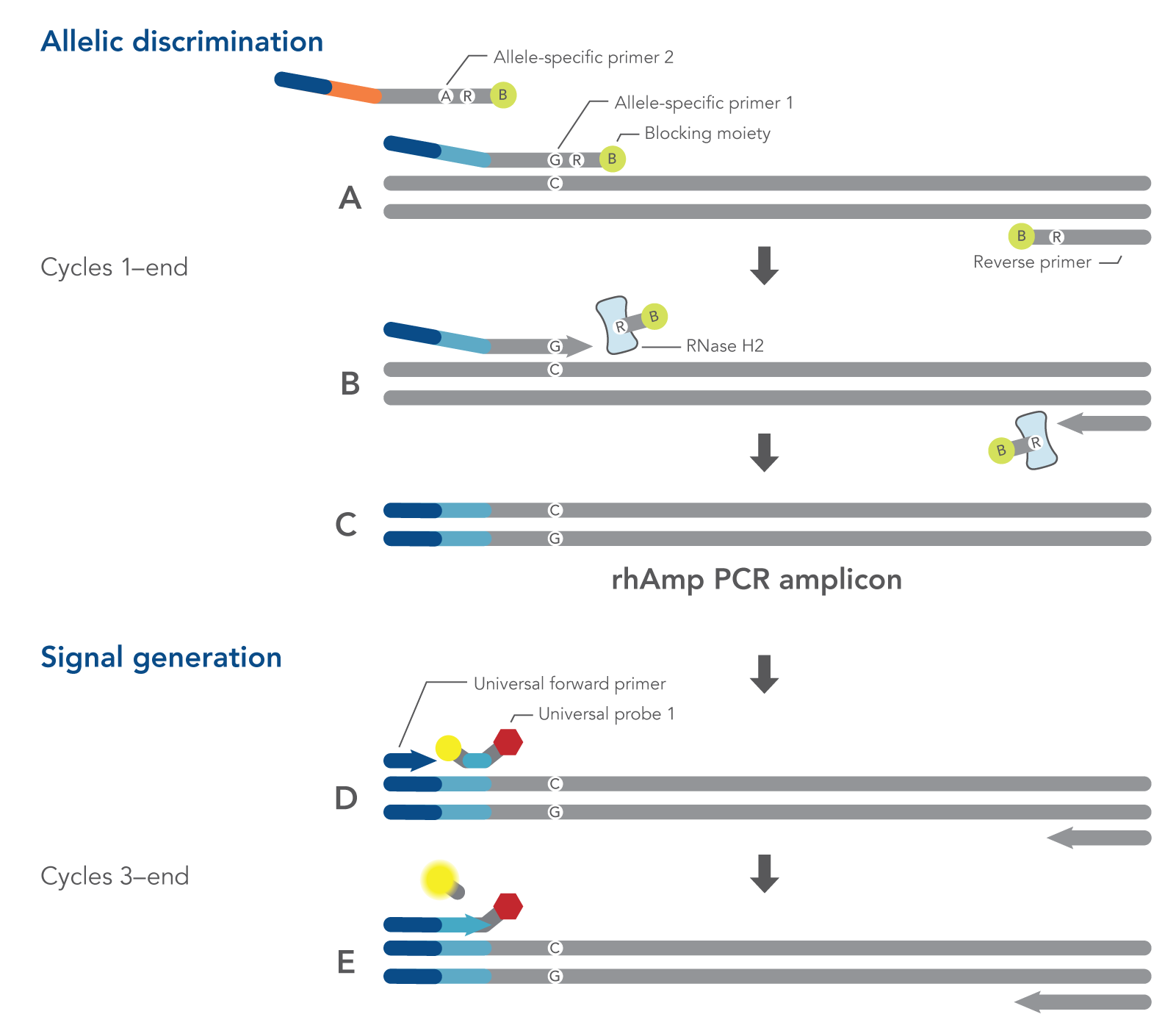
rhAmp SNP Genotyping is a quick, easy-to-use, single-tube process that allows for routine automation and delivers genotypes after only 90 minutes of PCR cycling time (Figure 1). rhAmp SNP Genotyping can be performed directly on a real-time PCR instrument or in a benchtop thermal cycler and transferred to a fluorescence detection platform for end-point analysis.
- rhAmp technology delivers enhanced precision and amplification without primer-dimers
- Novel dual-enzyme system increases mismatch discrimination
- Universal probe reporter system enables low cost per genotype
Figure 1. Schematic representation of a rhAmp SNP Genotyping PCR cycle. All components needed to measure both alleles are combined in a single reaction before cycling. (A) Both allele-specific primers query the SNP locus. (B-C) RNase H2 enzyme cleaves the primers that are hybridized to the perfectly matched target sequence, removing the RNA base and 3′ blocking modification, which allows extension by the Taq Polymerase. (A-C) During the first two amplification cycles, a tail sequence is incorporated into the amplicon that is subsequently recognized by (D) a universal probe-based reporter system. (E) Polymerase extension leads to degradation of the probe and signal generation.
rhAmp SNP Assays
The rhAmp SNP Assay library contains designs for human SNPs with a minor allele frequency >1%. The database currently contains >10 million predesigned SNP assays and continues to grow. rhAmp SNP Assays demonstrate consistently reliable results with >99.5% call accuracy in over 90% of tested assays. All human predesigned primer and probe sets are guaranteed to work, or IDT will replace them with a newly designed assay at no additional charge.
rhAmp ADME SNP Assays
rhAmp ADME SNP Assays target SNPs in human genes that are responsible for the absorption, distribution, metabolism, and excretion (ADME) of pharmaceutical compounds. These target SNPs are important biomarkers in pharmacogenetic studies. Each rhAmp ADME SNP Assay is experimentally tested to help ensure reliable data and signal generation.
Search for human rhAmp ADME SNP Assays »
Download an Excel file of available rhAmp ADME SNP Assays:
Custom rhAmp SNP Assays
The rhAmp Genotyping Design Tool can be used to design rhAmp SNP Assays for newly discovered SNPs that are not currently included in the predesigned human assay library, and for SNPs in any other species. The custom assay tool promotes design flexibility by accommodating short amplicons (as short as 40 bp).
For newly discovered human SNPs that are not currently in the predesign database, the design algorithm will perform BLAST and other sequence analyses to generate a suitable design. Custom design is easily performed by submitting target SNP sequences for any genome in FASTA format.
Design custom rhAmp SNP Assays
- All rhAmp SNP Assays are manufactured on demand and consist of 2 allele-specific forward primers and a locus-specific reverse primer
- Each assay is available in 2 mL tubes or matrix racks (96-well format). Assays are formulated and normalized to 20X or 80X concentration in IDTE pH 7.5 (1X TE Solution)
Table 1. Assay configurations.
| Product | Package size | Reactions* | Concentration |
|---|---|---|---|
| XS | 100 | 20X |
| S | 750 | 20X | |
| M | 2500 | 80X | |
| L | 6000 | 80X |
* Reaction size is for a 10 μL reaction volume.
Product Data
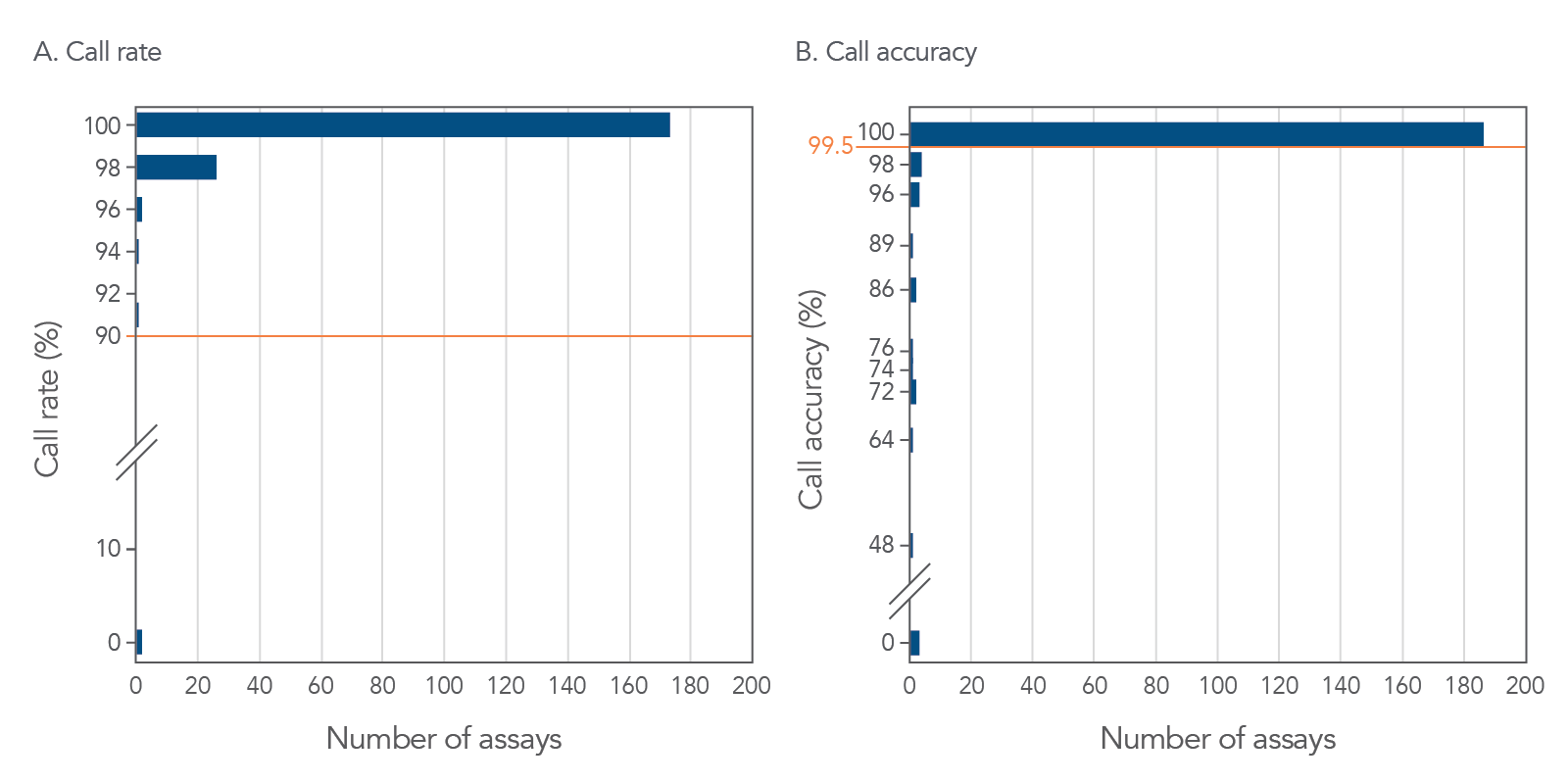
rhAmp SNP Assays provide excellent genotyping call rate and accuracy
More than 90% of rhAmp SNP Assays provide a ≥90% call rate and ≥99.5% call accuracy for reliable data generation in your genotyping experiments (Figure 2).
Figure 2. More than 90% of rhAmp SNP Assays tested provide >90% call rate and >99.5% call accuracy. Genotyping was performed on 213 rhAmp SNP Assays in 5 µL reactions, using human gDNA (3 ng) from 46 individuals (Coriell Institute). Analysis was conducted using QuantStudio™ 7 Flex Real-Time PCR System software (Thermo Fisher Scientific™).
High quality genotyping performance using gDNA from whole blood
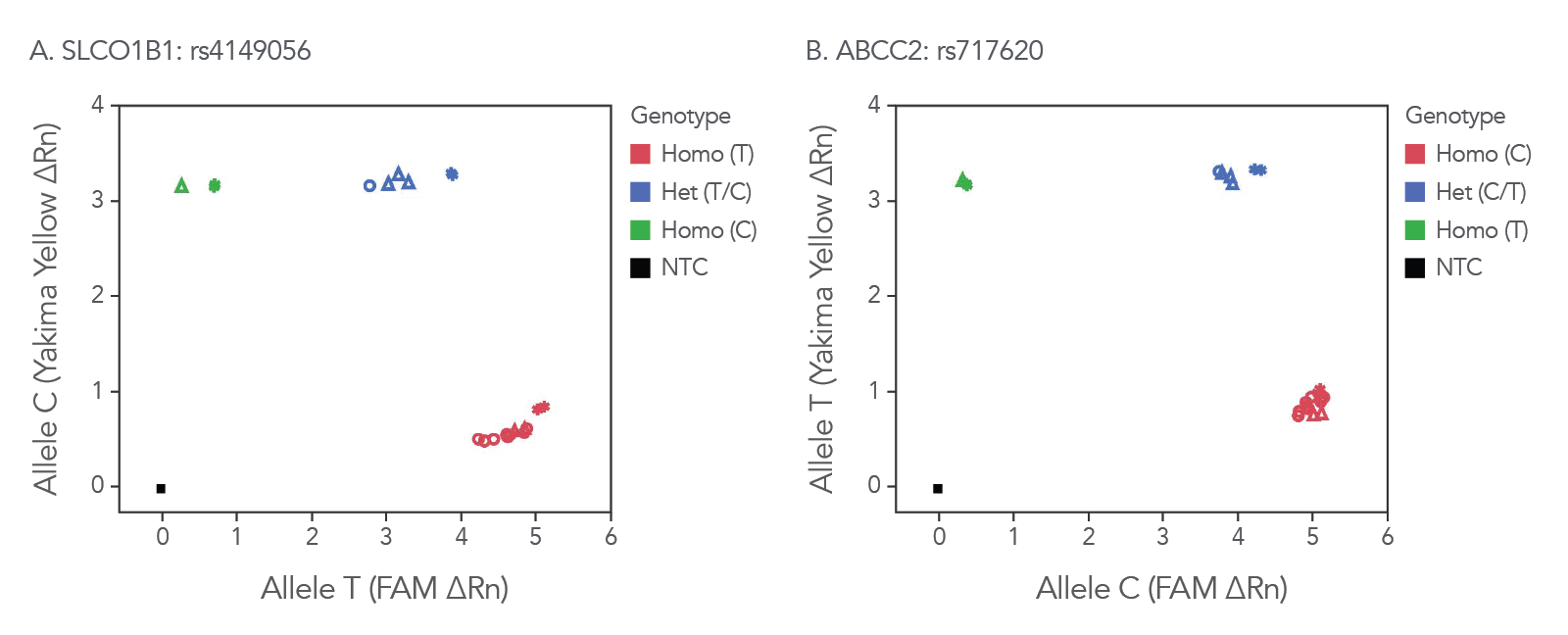
rhAmp SNP Genotyping offers reliable genotyping data across sample types. We have tested our assays and enzyme mixes across samples, including commercially available, purified genomic DNA and DNA isolated from whole blood that all perform similarly to synthetic gBlocks™ Gene Fragment templates (Figure 3).
Figure 3. rhAmp SNP Assays provide consistent results across sample types. Assays were designed for SNPs in genes (A) SLCO1B1 and (B) ABCC2. These assays were used to assess SNPs in 3 different sample types: synthetic gBlocks Gene Fragments (*), purified genomic DNA (Coriell Institute; ∆) and 6 replicates of genomic DNA isolated from whole human blood using the Qiagen QIAamp® DNA Blood Mini Kit (prepared by BioChain Institute; ○). Genotyping results were similar across sample types, as shown by overlapping symbols. Analysis was performed using QuantStudio™ 7 Flex Real-Time PCR System software (Thermo Fisher Scientific).
rhAmp primer technology reduces primer dimers and nonspecific amplification artifacts
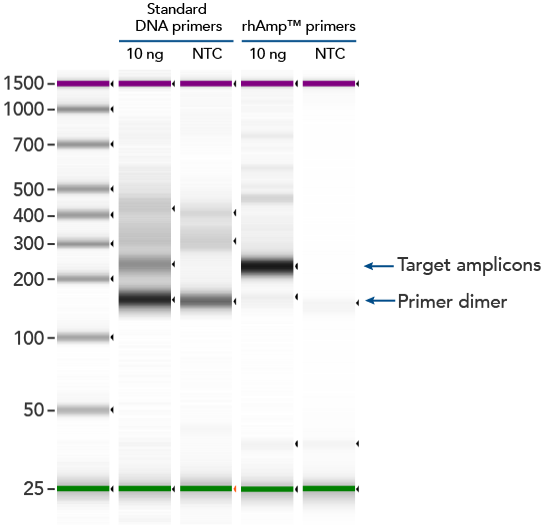
rhAmp SNP Genotyping technology uses activation of RNA-DNA hybrid primers by the RNase H2 enzyme from Pyrococcus abyssi. The primers are only activated upon precise annealing to the correct target, and this RNase H2 activation mechanism reduces primer-dimers and other amplification artifacts that can affect experimental results. Figure 4 shows how effective RNase H2 activation is for reducing primer-dimer artifacts, even when 96 primer pairs are multiplexed together in a single PCR experiment.
Figure 4. rhAmp primers reduce primer-dimers and nonspecific amplification artifacts in multiplex applications compared to standard primers. In multiplex PCR amplification of 96 targets from human genomic DNA [8 replicates of Coriell human gDNA sample (NA12878)], two sets of multiplex primers for the 96 targets (192 individual primers) were synthesized either as standard PCR primers or as RNase H2–activated, rhAmp primer pairs. The results show tight control of primer-dimer artifacts, even in highly multiplexed assays. Total reaction volumes were 50 µL, with 10 ng of genomic DNA template or no template controls. RNase H2 was present in all reactions but has no functional role in PCRs with standard primers.
Greater signal-to-noise ratio for higher confidence calls
rhAmp SNP Assays deliver an increased signal-to-noise ratio over traditional 5′-nuclease chemistry. In rhAmp technology, RNA-DNA hybrid primers are blocked at the 3′ end to prevent primer-dimer formation and non-specific amplification, mitigating the unbalanced consumption of reaction components. Increased signal-to-noise ratio provides improved cluster separation for higher confidence automated genotyping calls (Figures 5 and 6).
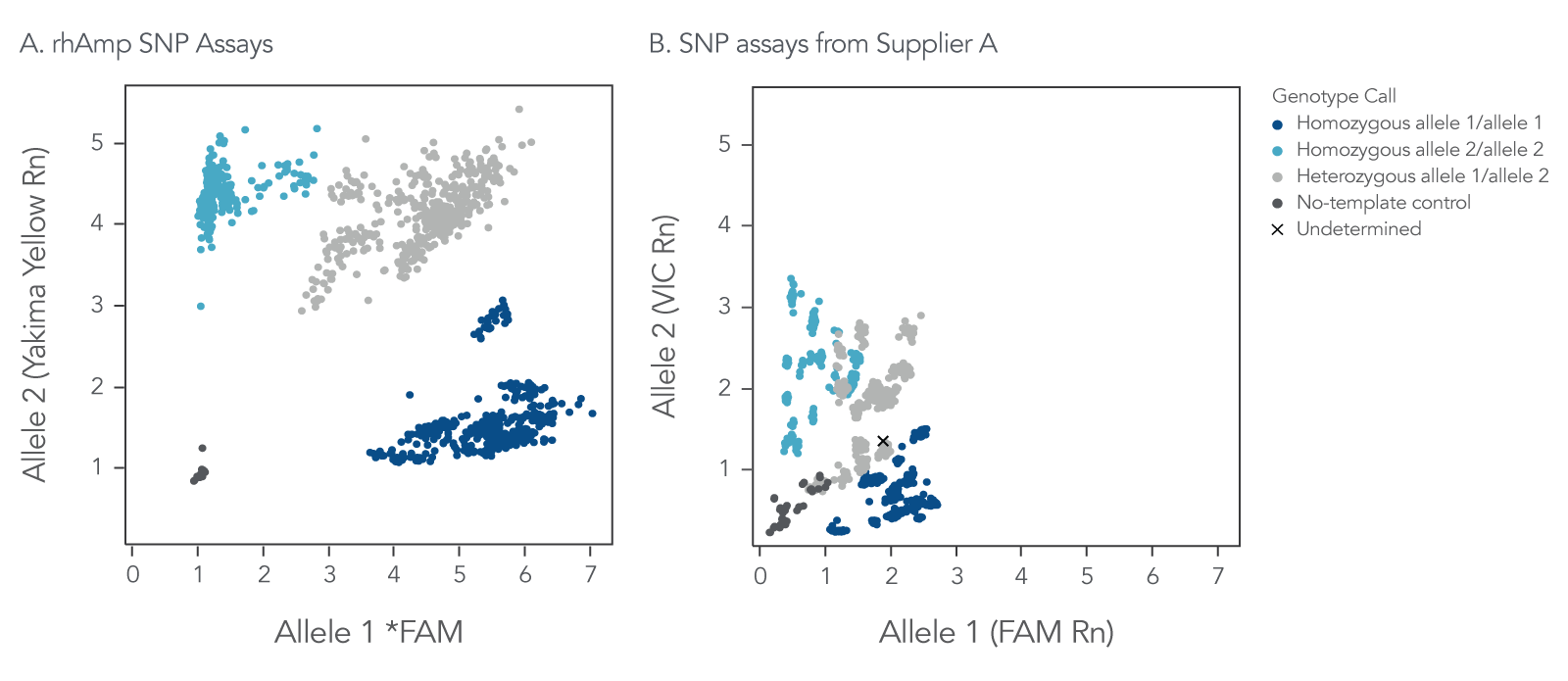
Figure 5. rhAmp SNP Assays deliver consistently higher genotype cluster signal and greater cluster angle separation in comparison to genotyping assays from Supplier A. rhAmp SNP Assays targeting 18 SNPs were evaluated against 5′-nuclease SNP assays from Supplier A targeting the same 18 SNPs. Genomic DNA samples (3 ng) from 46 individuals (Coriell Institute) were tested in 5 µL reactions, with all 3 possible genotypes represented. Two no-template, negative control (NTC) reactions were run for each assay. Post-PCR analysis of normalized reporter dyes (Rn) was used to generate allelic discrimination plots of (A) combined data for all 18 rhAmp SNP Assays, and (B) combined data for all 18 SNP assays from Supplier A. The universal reporter system used in rhAmp SNP genotyping contributes to the high normalized signal for all gDNA samples, the consistent signal from the negative control reactions, and the consistent cluster angles across all 18 SNP assays.
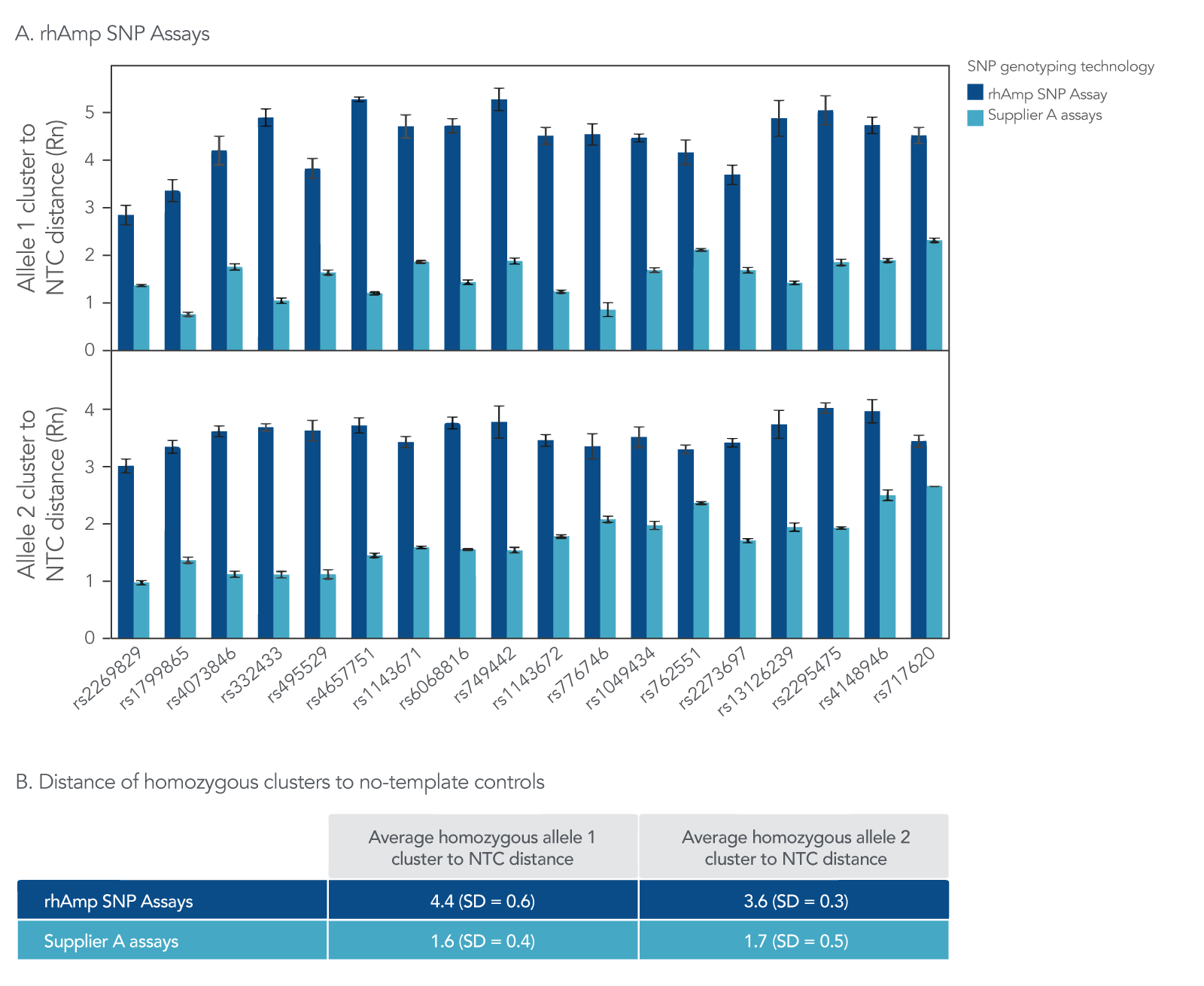
Figure 6. rhAmp SNP Assays result in higher average cluster to NTC distance than SNP assays from Supplier A. rhAmp SNP Assays targeting 18 SNPs were evaluated against 5’-nuclease SNP assays from Supplier A targeting the same 18 SNPs. Genomic DNA samples (3 ng) from 46 individuals (Coriell Institute) were tested in 5 µL reactions, with all 3 possible genotypes represented. (A) The average cluster to NTC distance describes the distance of a straight line between the homozygous cluster and NTC for each assay. The maximum average distance achieved for rhAmp SNP Assays was 5.3 for Allele 1 and 4.0 for Allele 2, while the maximum distance for Supplier A assays was 2.3 for Allele 1 and 2.7 for Allele 2. (B) The average homozygous cluster to NTC distance and standard deviation is shown across all 18 assays. The rhAmp SNP Genotyping technology consistently produced high cluster signal, with a 2.5-fold higher average cluster to NTC distance than assays from Supplier A.
rhAmp SNP Assays and reagents are made using the reliably synthesized oligonucleotides
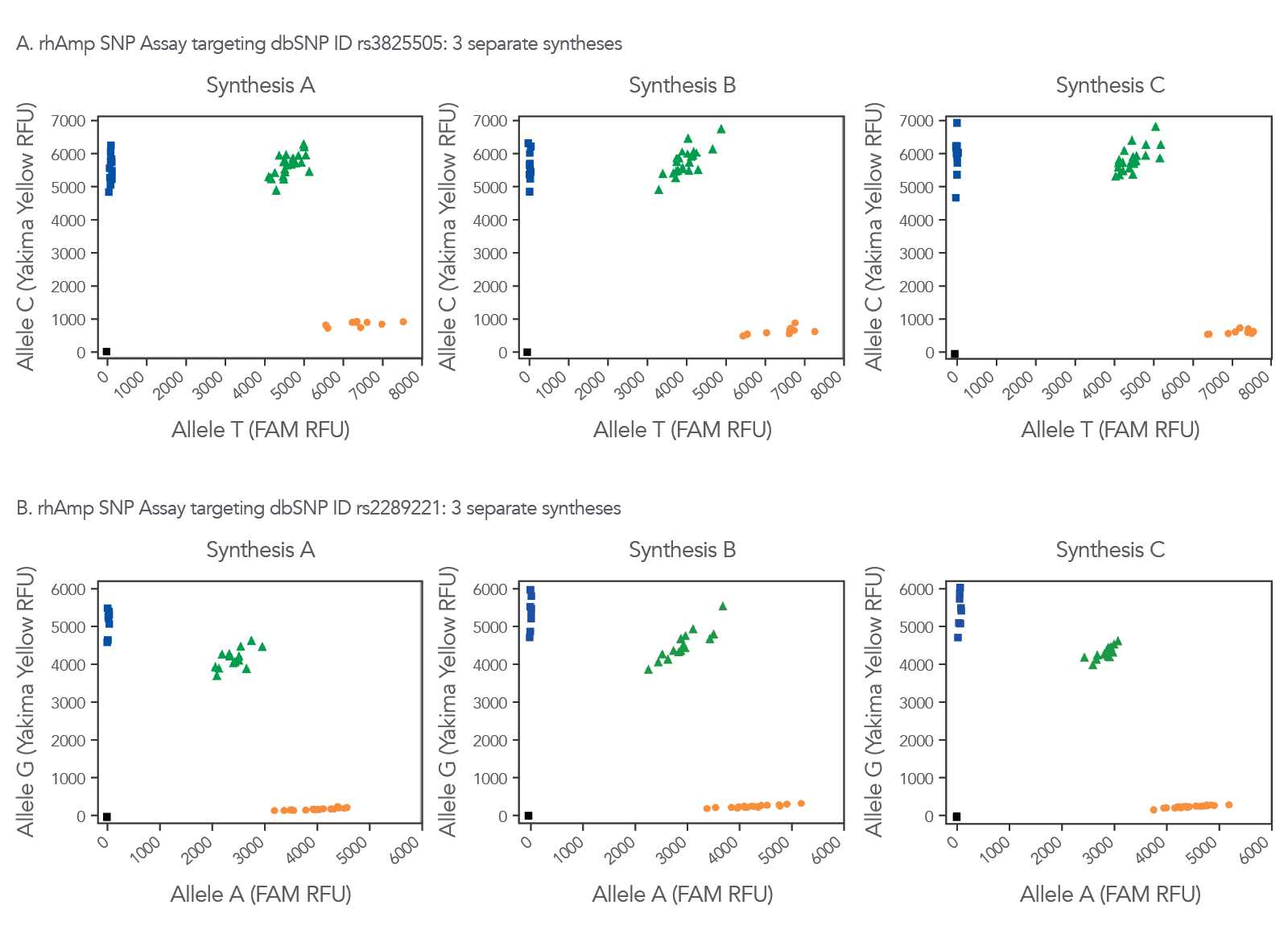
Consistent synthesis quality helps ensure that when you or another researcher orders a rhAmp SNP Assay for the same SNP ID, the results will not change (Figure 7).
Figure 7. Consistent synthesis quality helps ensure reliability of data with rhAmp SNP Assays. Three independent lots of rhAmp SNP Assays targeting SNP ID (A) rs3825505 and (B) rs2289221 were synthesized and formulated (Synthesis A, B, and C). Genotyping was performed in 5 µL reactions, using 3 ng human gDNA from 46 human gDNA samples (Coriell Institute). Data was analyzed using CFX Manager™ Software (Bio-Rad). Yakima Yellow™ signal was detected using the VIC™ channel without calibration.
rhAmp SNP Genotyping is compatible with commonly available real-time qPCR platforms
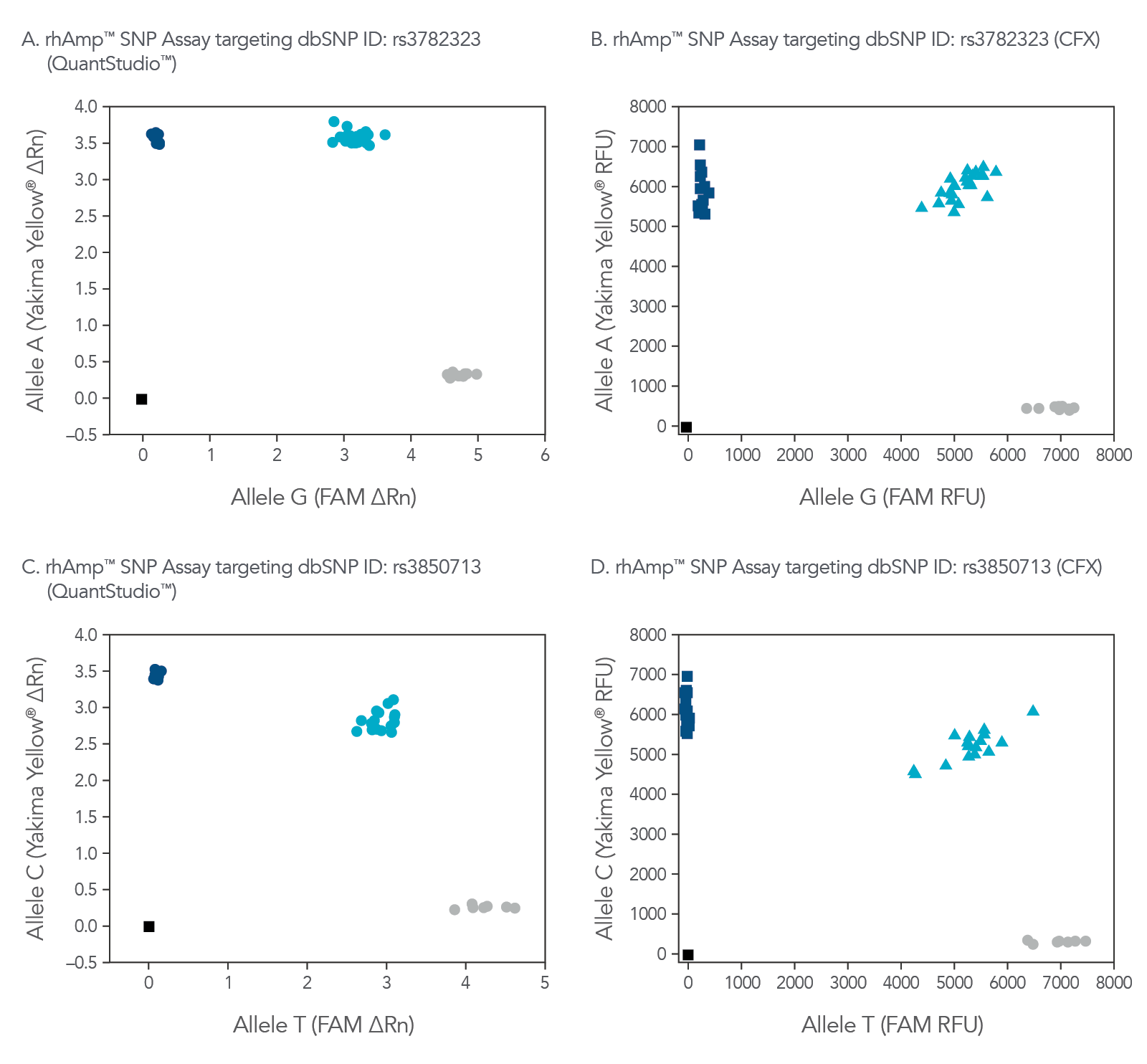
rhAmp SNP Assays are compatible with common real-time qPCR platforms (Figure 8). Universal probes are part of the rhAmp Reporter Mix, available with or without reference dye to ensure compatibility.
Figure 8. rhAmp SNP Assays deliver high precision regardless of qPCR instrument used. Allelic discrimination plots for SNPs located in genes (A and B) SMARCD1 (rs3782323) and (C and D) LINC00111 (rs3850713) show consistent results generated by rhAmp SNP Assays analyzed using QuantStudio™ Real-Time PCR software (Thermo Fisher Scientific) or the CFX Real-Time PCR software (Bio-Rad). Genotyping was performed in 5 µL reactions using 3 ng gDNA from 46 human gDNA samples (Coriell Institute). Yakima Yellow™ signal was detected using the VIC™ channel without calibration. More than 90% of rhAmp SNP Assays demonstrate a call rate >95% and call accuracy of 99.5% regardless of qPCR instrument.