SARS-CoV-2 sequencing
When it mattered most, xGen COVID-19 NGS solutions enabled researchers to sequence the SARS-CoV-2 genome and its variants. As the era of genomics medicine takes hold, you can count on xGen NGS to continue providing innovative solutions for viral variant identification, tracking, and surveillance.
xGen™ NGS—made for COVID-19 research.
Overview
- Recommended products—curated for SARS-CoV-2 sequencing
- A tested solution—takes you from research sample to sequencer
- Comprehensive coverage—amplicon panel provides up to 99.7% genome coverage of the major SARS-CoV-2 variants
- Save time, reduce costs—cDNA to sequencer in approximately 3 hours
- Flexible sample input—compatible with 1st or 2nd strand cDNA synthesis and low viral loads from a variety of sample types
- Streamlined workflow—driven by xGen Amplicon technology that generates sequencing libraries in a single tube
- Automation friendly—easy workflow for the 96-reaction size
Viral genome sequencing of SARS-CoV-2
Next generation sequencing (NGS) approaches have proven to be an invaluable tool for understanding the coronavirus responsible for the global COVID‑19 pandemic, SARS-CoV-2 [1]. Viral sequencing of the SARS-CoV-2 genome helps researchers identify novel variants of the virus so their response to this public health crisis can be data-driven and timely.
Sequencing SARS-CoV-2 with the xGen Amplicon workflow
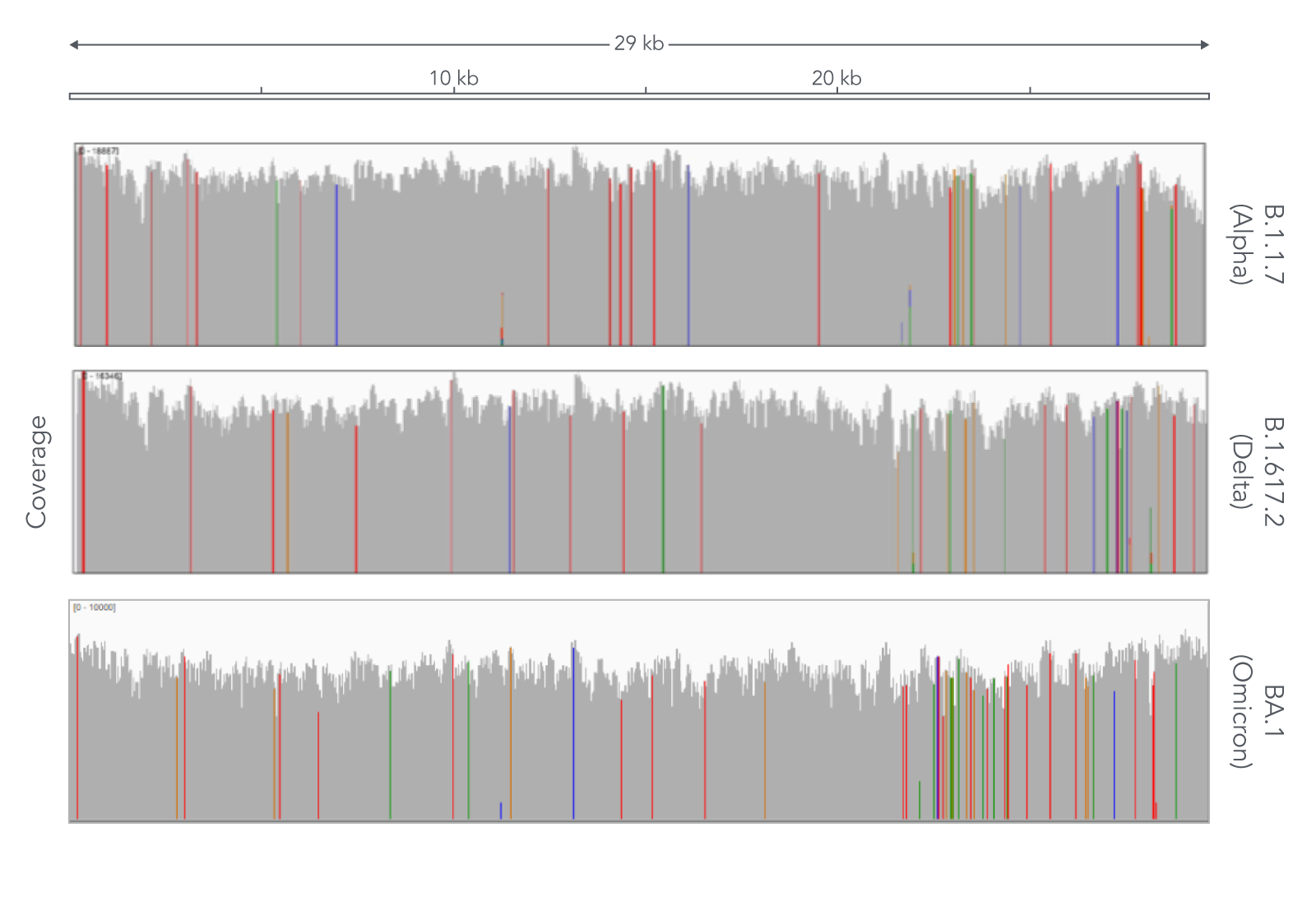
Due to the speed with which variants of SARS-CoV-2 have appeared, researchers have had to repeatedly change methods to effectively track SARS-CoV-2 variants. Despite the ever-emerging variants of SARS-CoV-2, the xGen SARS-CoV-2 Amplicon Panel has not resulted in coverage dropouts of the SARS-CoV-2 genome in any of the variants that we’ve tested (Figure 1). This high genome coverage is maintained by xGen Amplicon technology’s ability to generate super amplicons which can provide comprehensive coverage in regions that have mutations in primers sites for shorter amplicons. This means that the xGen SARS-CoV-2 Amplicon Panel has helped researchers to comprehensively identify variants and improve their understanding of the virus’ evolution, transmission, and pathogenesis.
The xGen Amplicon workflow for SARS-CoV-2 sequencing is streamlined and cost-effective and includes the xGen Amplicon Core Kit, the xGen SARS-CoV-2 Amplicon Panel, and the xGen Amplicon UDI Primers. These products have been selected for researchers using whole genome viral sequencing of SARS-CoV-2 as a method of variant tracking.
The xGen Amplicon Core Kit is a targeted amplicon library preparation kit for contiguous genome coverage (overlapping amplicons). This kit, along with the xGen SARS-CoV-2 Amplicon Panel, gives researchers the ability to sequence emerging variants of SARS-CoV-2 from a variety of samples such as nasopharyngeal/oropharyngeal swabs, sputa, or wastewater without needing to evaluate new primer sets.
The xGen SARS-CoV-2 Amplicon Panel uses overlapping amplicons to generate 345 amplicons, sized 116–255 bp (average 150 bp), along the length of the 29.9 kb viral genome, obtaining 99.7% coverage of the SARS-CoV-2 genome (Figure 1). This product is driven by the xGen Amplicon technology that generates sequencing libraries in a single tube, helping researchers go from cDNA to ready-to-sequence libraries faster and more efficiently. Furthermore, the single tube approach generates super amplicons that span across two intended amplicons, making them longer and providing researchers with additional confidence and coverage for variant calling. For further information about the xGen SARS-CoV-2 Amplicon Panel, read the DECODED article, Viral surveillance using wastewater.
Finally, the use of the xGen Amplicon UDI Primers further streamlines this process for multiplexing applications. The premixed primers condition the libraries for Normalase™ technology which uses a proprietary enzymatic library normalization procedure. The Normalase™ technology enables streamlined library balancing and pooling process without the need to quantify samples and maximizes sequencing efficiency.
IDT also offers support for downstream analysis of data generated with this workflow. Please refer to the technical note, xGen SARS-CoV-2 Amplicon Panel dockerized data analysis for more information.
Extraction
Reverse transcription
Sequencing & analysis
Method data
Comprehensive coverage of three major SARS-CoV-2 variants
To assess the xGen SARS-CoV-2 Amplicon Panel, RNA was extracted from nasopharyngeal or oropharyngeal swabs and converted into cDNA using the SuperScript IV Kit (Thermo Fisher Scientific). The resulting library generated with the xGen SARS-CoV-2 Amplicon Panel was sequenced 2 x 150 bp on a MiniSeq System (Illumina®). Resulting reads were subsampled to 83,000 reads per sample for analysis.
Across all three variants, the xGen SARS-CoV-2 Amplicon Panel provided comprehensive viral genome coverage (99.7%, on average), meaning that researchers have been able to use this panel to enrich and sequence the genome of SARS-CoV-2 throughout the pandemic without needing to re-evaluate new primers each time a new variant emerged (Figure 1).
More information about the effectiveness of the xGen SARS-CoV-2 Amplicon Panel can be found in the DECODED article, Viral surveillance using wastewater.
For further reading on SARS-CoV-2 viral genome sequencing analysis, see the technical note xGen SARS-CoV-2 Amplicon Panel dockerized data analysis guidelines.
Ordering
Resources
References
1. Singh D, Yi SV. On the origin and evolution of SARS-CoV-2. Exp Mol Med. 2021;53(4):537-547.